

If this does not occur, repeat the process with the reverse complement sequence file in a New alignment. I paste these into Microsoft Word and use search and replace to get rid of extra details. Repeat this process for the pstblue1vector. This file contains the sequence of the multiple cloning site region of pSTBlue Select from the next next residue to the end. For each gene within a dataset I usually have this file with the forward, reverse and consensus. I usually import all the forwards and reverses into a new BioEdit file.
Bioedit alignment tutorial password#
Each group should log on to a PC using the class ID bisc and the password pseud. Depending on how well your reverse sequences overlap with your forwards, scroll right until they overlap with good sequences.Ĭlick on the edited forward sequence file to open it.
Bioedit alignment tutorial full#
See sequence analysis references for full map. Select the N and replace it by typing in the appropriate base. Now scroll right again and look for any bases that need checking. The regular copy and paste features work between copies of the program, but copying and pasting sequences does not. I select a point in the reverse, then select sequence to the end Edit, Select to End, control-e. This changes the way the sequences are displayed. This creates a duplicate sequence that can be edited without changing the original sequence.Įditing and most of the analysis will be done using BioEdit, a freeware sequence analysis program developed by Tom Hall at North Carolina State University. All of that probably sounds very confusing, once you have carefully worked through it a couple of times it becomes very easy. I then select those sequences control-shift-acut control-shift-c or copy them control-a and paste them control-s to the desired BioEdit file. Once I am happy with that I ready to create what will become the consensus sequences. Return to your edited forward sequence file, delete the vector sequences, and save for next week. To get the sequence of the original template strand, the Reverse Complement must be prepared.Ĭreate a new BioEdit file. Select all the reverse sequences and cut them. Change the view type on the lower toolbar 3rd of the alignment windowselect the third colored button from the left says Shade identities and similarities when you hold the mouse over it. Click on Sequence menu, Tutorlal alignmentAlign two sequence allow ends to slide.Īlso copy the file pstblue1vector.

Then I run a NJ analysis to see what is going on with the dataset. Guide to editing sequences with Chromas and BioEdit
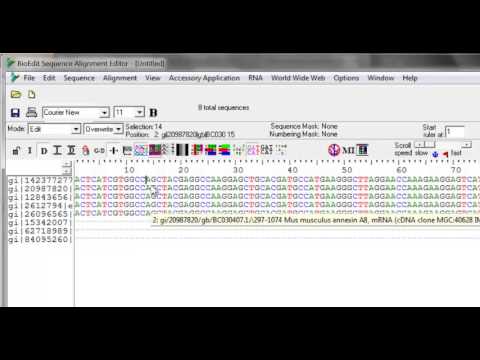
I copy the sequence titles to the clipboard Edit, Copy sequence titles. I manually align them and check for obvious missing bases and either correct them or add a gap to preserve the alignment. Click on the view menu for the original unedited fileand check Reverse Complement.

Select the value you wish to change, hit the value on the keyboard and that will reset it. Overwrite the sequence title onto the next title shifting up, when the title is being edited. MEGA also has an alignment editor, but I’ve not really used it very much. BioEdit can also edit chromatograms, but I find Chromas to be nicer. BioEdit is a mouse-driven, easy-to-use sequence alignment editor and sequence analysis program designed and written by a graduate student. This is likely to be the final release of BioEdit. North Carolina State University, Department of Microbiology.


 0 kommentar(er)
0 kommentar(er)
